Rees EJ, Erdelyi M, Kaminski-Schierle GS, Knight AE and Kaminski CF, "Elements of image processing in localisation microscopy", Journal of Optics 15, 094012 (2013). DOI |
Localization microscopy software generally contains three elements: a localization algorithm to determine fluorophore positions on a specimen, a quality control method to exclude imprecise localizations, and a visualization technique to reconstruct an image of the specimen.
Such algorithms may be designed for either sparse or partially overlapping (dense) fluorescence image data, and making a suitable choice of software depends on whether an experiment calls for simplicity and resolution (favouring sparse methods), or for rapid data acquisition and time resolution (requiring dense methods). We discuss the factors involved in this choice. We provide a full set of MATLAB routines as a guide to localization image processing, and demonstrate the usefulness of image simulations as a guide to the potential
artefacts that can arise when processing over-dense experimental fluorescence images with a sparse localization algorithm.
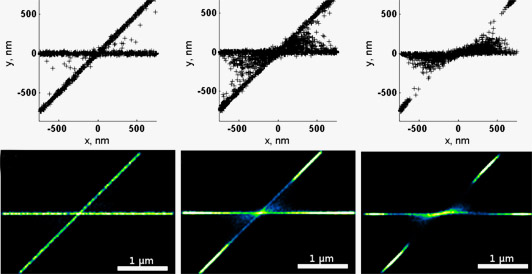 Effect of single molecule positional errors on image reconstructions. The data model dye molecules along two lines, a situation encountered for example in the superresolution imaging of two crossing protein fibrils. From left to right the number of mislocalisations increases. Notice that localistion errors are most likely to occur in regions between the two fibrils, near their crossing point. Notice also that the quality control mechanism 'rejects' several molecules on the two fibrils near their crossing point (blue or missing regions).
Effect of single molecule positional errors on image reconstructions. The data model dye molecules along two lines, a situation encountered for example in the superresolution imaging of two crossing protein fibrils. From left to right the number of mislocalisations increases. Notice that localistion errors are most likely to occur in regions between the two fibrils, near their crossing point. Notice also that the quality control mechanism 'rejects' several molecules on the two fibrils near their crossing point (blue or missing regions).
