New modes of optical microscopy offer insights on the nanometre scale, combining the advantages of traditional high resolution methods, such as electron microscopy, with the advantages of using light for probing life and matter.
We develop the whole gamut of advanced microscopy techniques in the group and are thus able to respond flexibly to problems in scientific imaging. This article focuses on optical superresolution techniques we develop, often in collaboration with leading researchers from around the world. Optical superresolution imaging refers to the recovery of spatial information on the sub wavelength scale, below the limit imposed by optical diffraction.
Much of what we do in this area is motivated by challenging demands from biological research where processes occur over a huge range of temporal and spatial scales. For example, the interaction of two proteins may occur on the microsecond time-, and nanometre spatial-, scales, whereas the development of an organism from a few cells to a functioning system can extend over days in time and millimetres in space. No single imaging modality can serve all questions: Our strategy is to pick what works best or otherwise to develop new and better techniques. We are interested also in applications outside of biology for example in condensed matter physics (e.g. basic light - matter interactions) and the design of functional nanomaterials.
If you have an interest in collaborating with us in this field, we would be pleased to hear from you. Superresolution techniques we develop include dSTORM, SIM, mSIM, RESOLFT and STED) and we present these below.
dSTORM
Direct stochastic optical reconstruction microscopy is a class of super-resolution techniques that operates on the principle of single molecule localisation imaging. Through the use of photoswitchable dyes and imaging molecules one by one, it is possible to locate their spatial position within a sample with a resolution of about one fiftieth of the wavelength of light (ca10 nm). We have developed a setup that permits us to resolve two colours and we are flexible to use most of the dyes commonly used in superresolution microscopy. We develop algorithms to improve localisation precision and speed [1,2,3] and constantly improve and refine experimental strategies in this area. We collaborate strongly here with the National Physics Laboratory and the Sauer group in Würzburg.
Our dSTORM microscope platform serves equally well for use in other single molecule and high resolution imaging modalities such as PALM, SOFI etc. and is currently lead by Romain Laine.
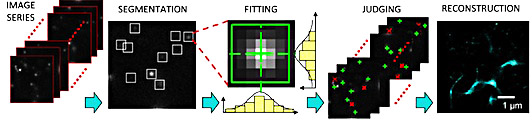
Figure 1. Elements of image processing in dSTORM imaging [1]
Applications of dSTORM
Neurodegeneration. We have pioneered the use of dSTORM for the nanoscale imaging of neurotoxic proteins [4] and use it in the study of prion like propagation of misfolded protein species [5]. We have recently used 2 colour dSTORM to visualise directly, at the molecular level, how misfolded species evolve in time [6]. These results provide unprecedented mechanistic insights into the molecular origins of disesases such as Alzheimer's and Parkinson's. This research is strongly collaborative with the Neurodegenerative Disease Consortium, led by Prof. Peter St. George Hyslop.
Virus particle assembly. In collaborations with the laboratories of Dr. Colin Crump we study the molecular structure and function of proteins involved in the formation of virus particles, central to a range of infectious diseases. This study is a close collaborative work with Dr Colin Crump (Virology department, Addenbrooke’s hospital) and Dr. Sebastian van de Linde from Würzburg.

Figure 2. Schematic makup of an HSV1 virus particle. We use dSTORM imaging to reveal the spatial organisation of proteins throughout the virus.
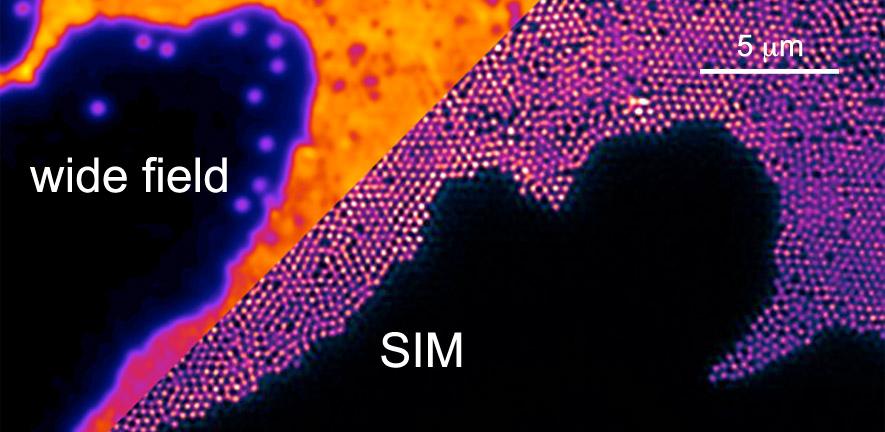
Structured Illumination Microscopy (SIM)
This technique offers spatial resolution which is twice as high as for confocal imaging in each dimension and it is compatible for use with all conventional fluorophores used in biology. The technique offers superresolution by projecting a structured illumination pattern onto the sample. Spatial frequencies in the sample and illumination patterns mix and this shifts high frequency information from the sample into the lower frequency pass band of the optical imaging system; in other words information can now be recovered from the sample that would not otherwise be visible. The beauty of SIM is the flexibility with which it can be deployed. We use supercontinuum lasers for multicolour excitation and spatial light modulators to project the patterns. This permits us to acquire images in fractions of a second.
A related technique is mSIM (multifocal SIM) which combines aspects of SIM and confocal imaging, offering higher resolution than confocal microscopy and better imaging depth than what is possible via SIM.
Applications of SIM and mSIM
The SIM was developed by Laurie Young and we use it in a variety of projects ranging from live cell imaging to watching the growth of proteins into fibrillar aggregates in real time. mSIM is also suitable for imaging at depth for example in thicker biological probes, such as tissue from brains or organisms. The instrument is flexibly configured and can be switched between SIM and mSIM imaging modes at the switch of a button. We develop our own algorithms and software to reconstruct superresolution images from SIM and mSIM images. We colllaborate with Dr. Kevin o'Holleran and the group of Prof. Rainer Heintzmann in Jena.

Figure 3. mSIM illumination pattern generated with a spatial light modulator. The technique has similarities in principle to a confocal microscope with multiple illumination beams
Stimulated Emission Depletion Microscopy (STED)
Principle
STED microscopy [7] shares principles of confocal laser scanning microscopy and structured illumination microscopy. In traditional confocal microscopy a laser beam is focused to a diffraction limited spot, referred to as the point spread function, PSF. The PSF has a diameter of ca 250 nm and excites fluorescence in the sample, which is collected as the beam is raster-scanned across the region of interest (Figure 4a). One can think of the laser beam as a paint brush - the thinner the tip, the finer the detail we can recover from the sample ('Excitation beam' shown in blue in figure below). The trick of STED, or more generally the RESOLFT principle, is to 'shape' the tip into a spot much smaller than the 250 nm diffraction limit, by using a so-called depletion beam, which has a doughnut shaped intensity distribution, with zero intensity in the middle and high intensity in an annular region around the central position ('STED beam' in the figure below). Superimposing this depletion beam on the excitation beam prevents spontaneous emission (fluorescence) from fluorophores anywhere except in a very small central region, thus yielding an effective PSF of only 30 to 90 nm in diameter (Figure 4b-c). This permits one to gain information on a much smaller scale than possible with standard confocal imaging. The technique works at a reasonable depth and is thus useful to look at distribution of molecules deep within cells.
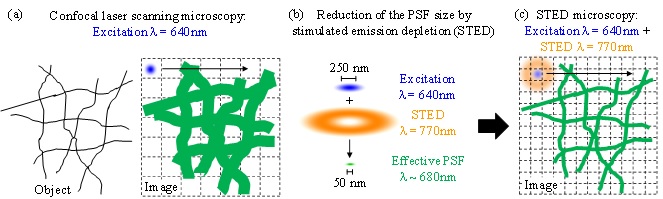
Figure 4. Principle of Stimulated Emission Depletion microscopy (STED)
Application of STED to study amyloid fibril formation
In work lead by Pierre Mahou we have built a super resolution optical microscope based on Stimulated Emission Depletion microscopy (STED) to study amyloid fibrils labelled with red dyes such as the ATTO or Aberrior dyes. The excitation of various fluorophores is enabled by a supercontinuum source generated by pumping a photonic crystal fiber and the STED beams is produced by a spatial light modulator that also corrects for the optical aberrations of the system. This microscope has been successfully evaluated with ideal test samples (Figure 5) and is now being used to monitor the formation of amyloid fibrils in vitro.
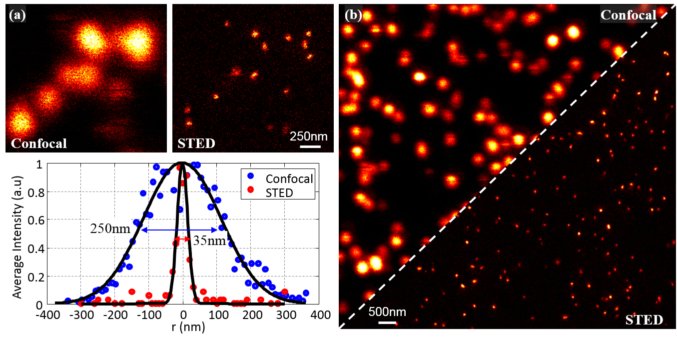
Figure 5. Evaluation of the performance of the lab-built STED microscope from ideal calibration samples. Representative images of 20 nm fluorescent beads taken in confocal and STED. Typically the resolution in the super-resolution mode is of the order 35 nm, which corresponds to an improvement by a factor 7.
Contacts
To discuss potential applications for super-resolution microscopy in your research and the use of any of our instruments please contact Edward Ward (ew535@cam.ac.uk). Alternatively, you can also contact the group leader Prof. Clemens Kaminski (cfk23@cam.ac.uk).
References
[1] Rees EJ et al. "Blind Assessment of Localisation Microscope Image Resolution", (2012).
[2] Erdelyi M et al. “Correcting chromatic offset in multicolor superresolution localization microscopy” (2013)
[3] Sinko J et al. "TestSTORM: Simulator for optimizing sample and image acquisition in localization based super-resolution microscopy" (2014)
[4] Kaminski Schierle GS et al. “In Situ Measurements of the Formation and Morphology of Intracellular β-Amyloid Fibrils by Super-Resolution Fluorescence Imaging” (2011)
[5] Michel CH et al. "Extracellular Monomeric Tau is Sufficient to Initiate the Spread of Tau Pathology" (2014)
[6] Pinotsi D et al. “Direct Observation of Heterogeneous Amyloid Fibril Growth Kinetics via Two-Color Super-Resolution Microscopy” (2013)
[7] Hell, S. W. et al. "Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy" (1994)
